
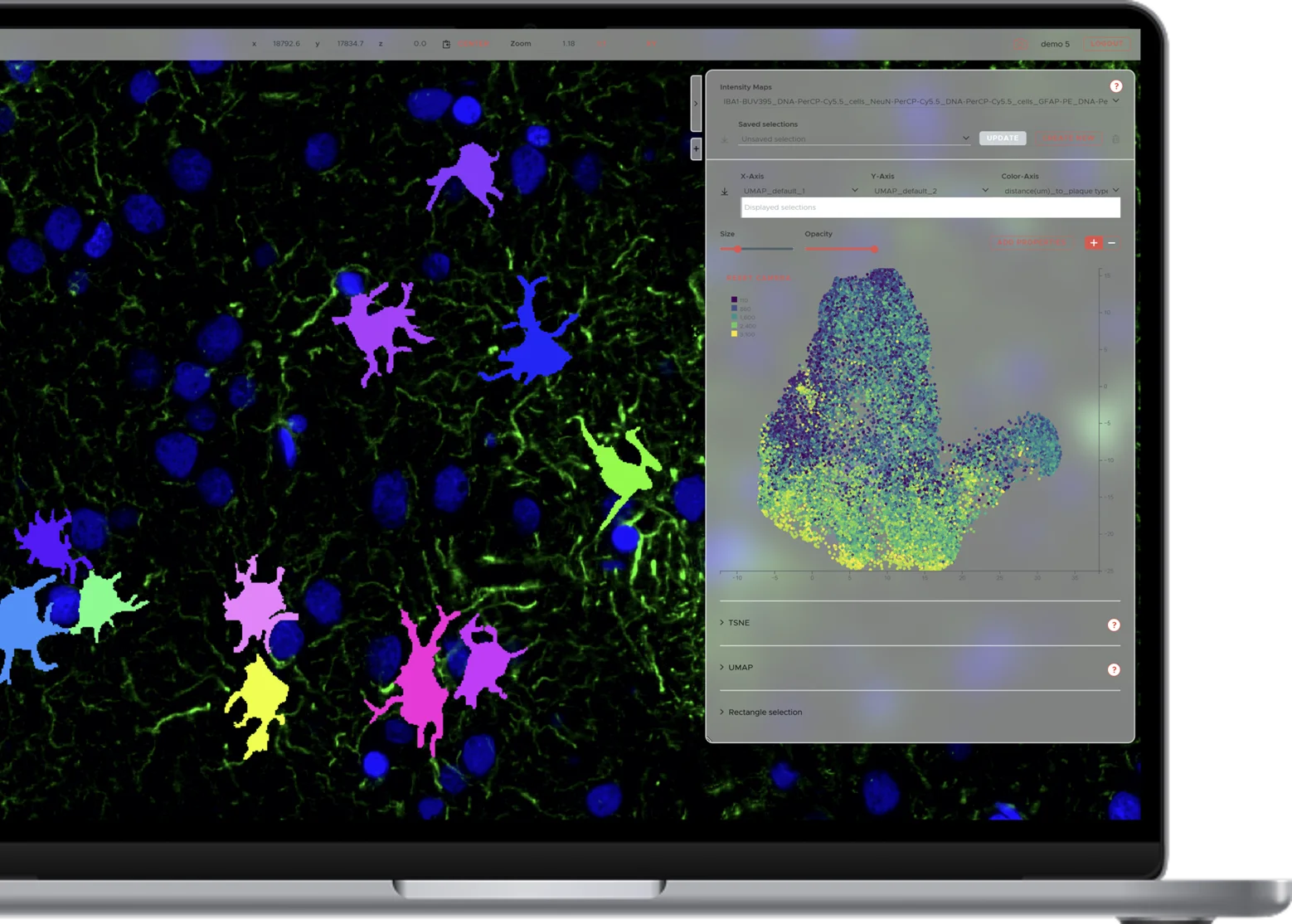
World-class cell segmentation.
Also for cells with complex morphologies such as neurons and glia. Supports CD45, NeuN, Iba1, GFAP, DAPI and many more.

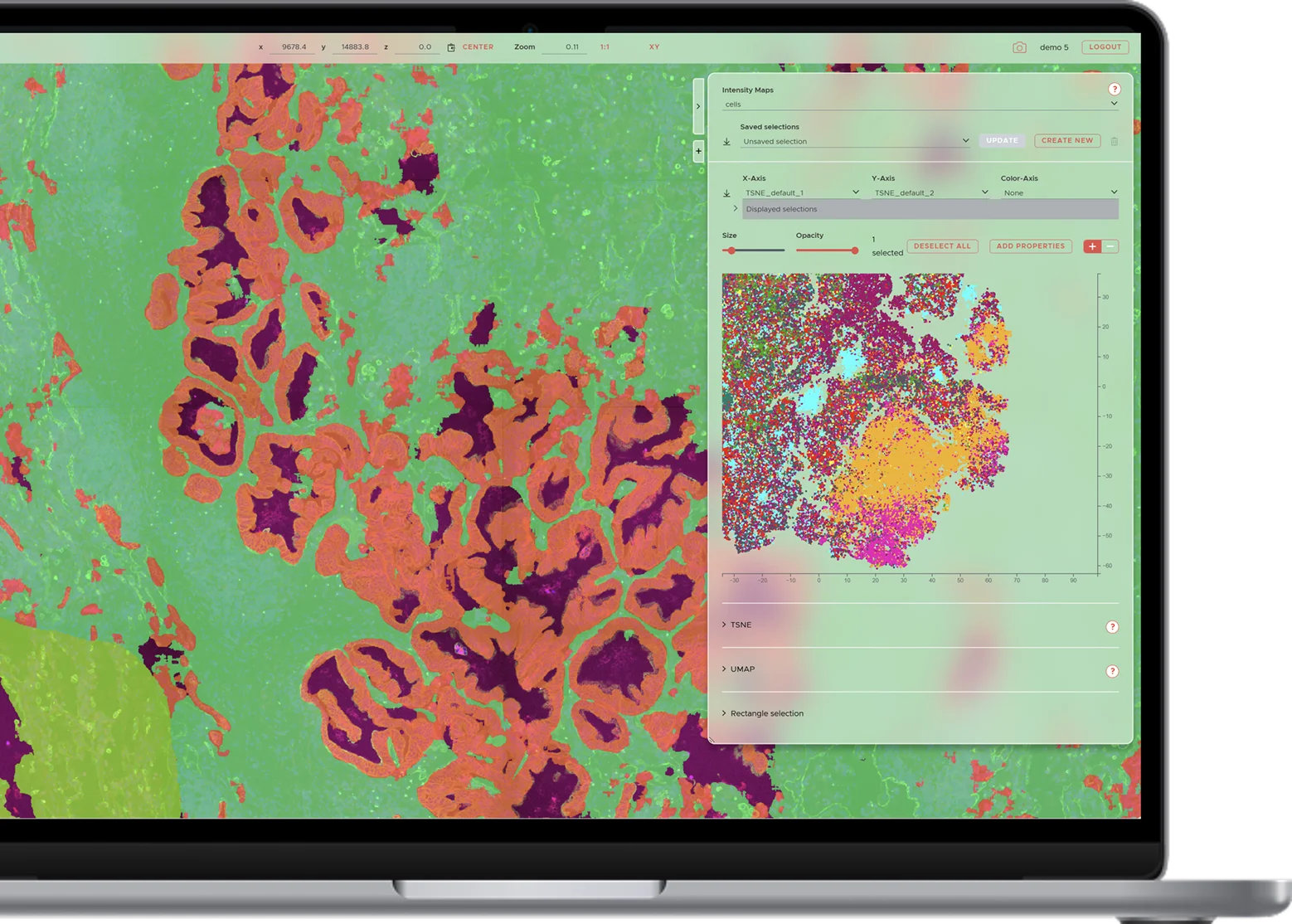
Neuropathology analysis.
Classify pathological protein aggregates of Amyloid β, pTau, α-Synuclein, TDP43 and more.
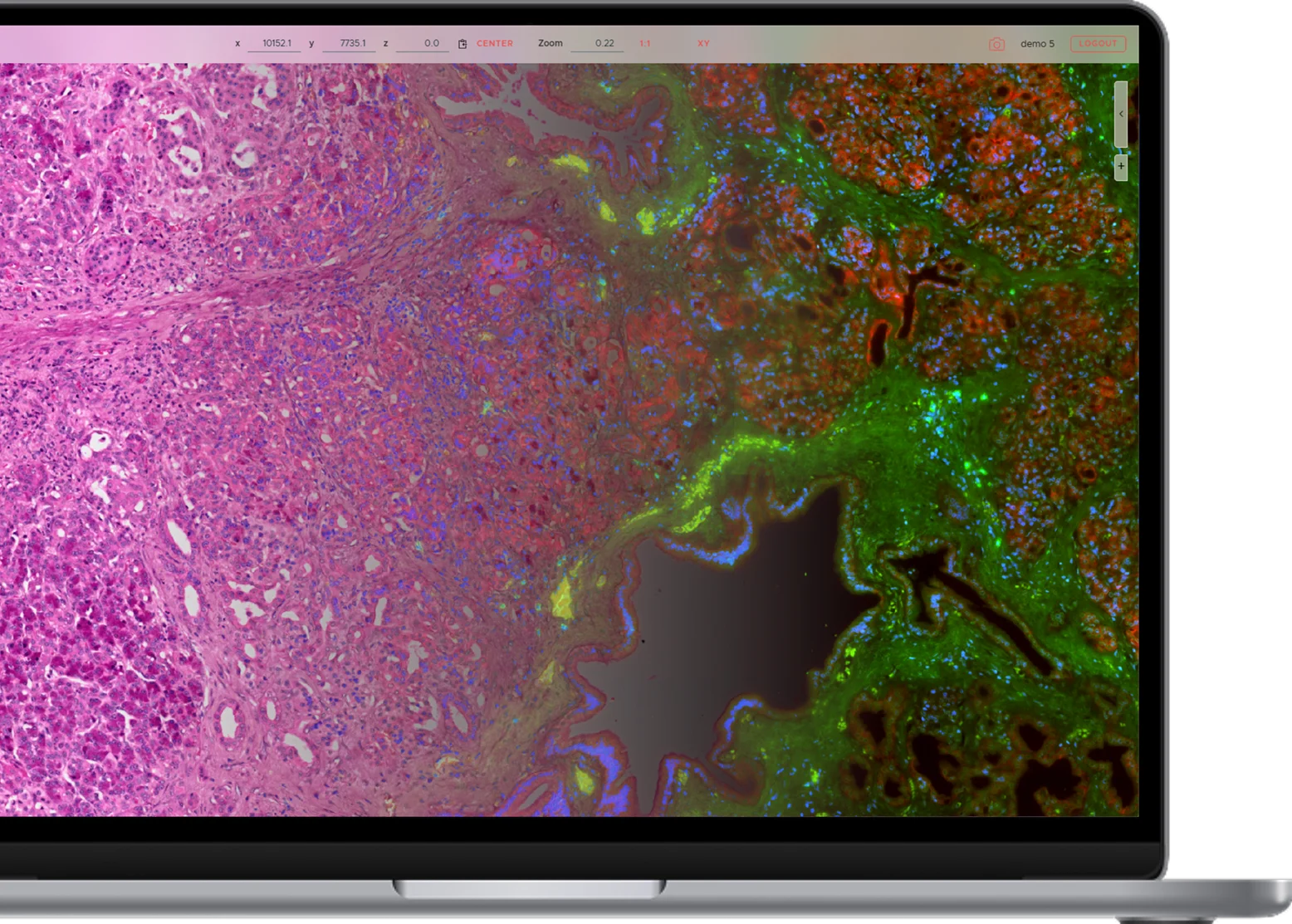
Segment H&E images.
Leverage well-established IHC protocols, multiplexed.
Elastic registration.
Correct complex deformations accross consecutive staining cycles, protocols or physical sections.





Workflow
SPATIALTM provides access to an end-to-end image analysis workflow — from data upload to spatial biomarker discovery.
Start Now
applications
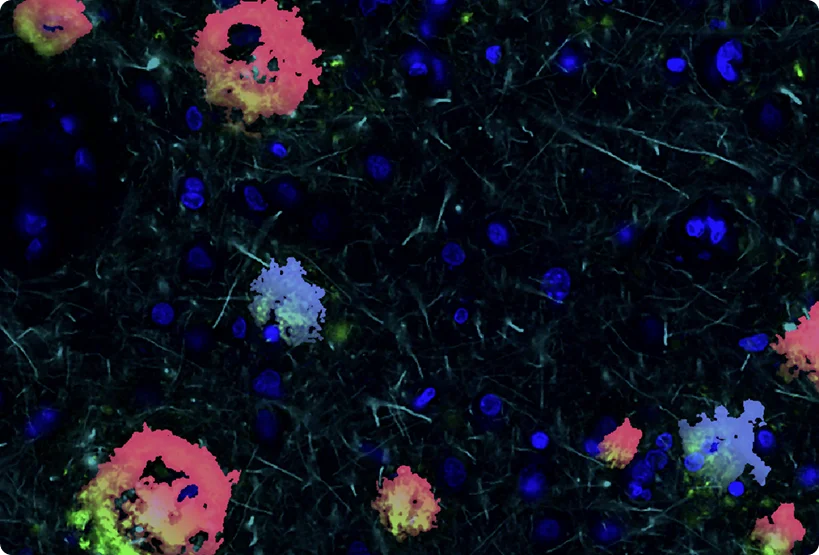
Comprehensive in silico tissue characterization in Alzheimer's and Parkinson's disease by spatial proteomics
Learn more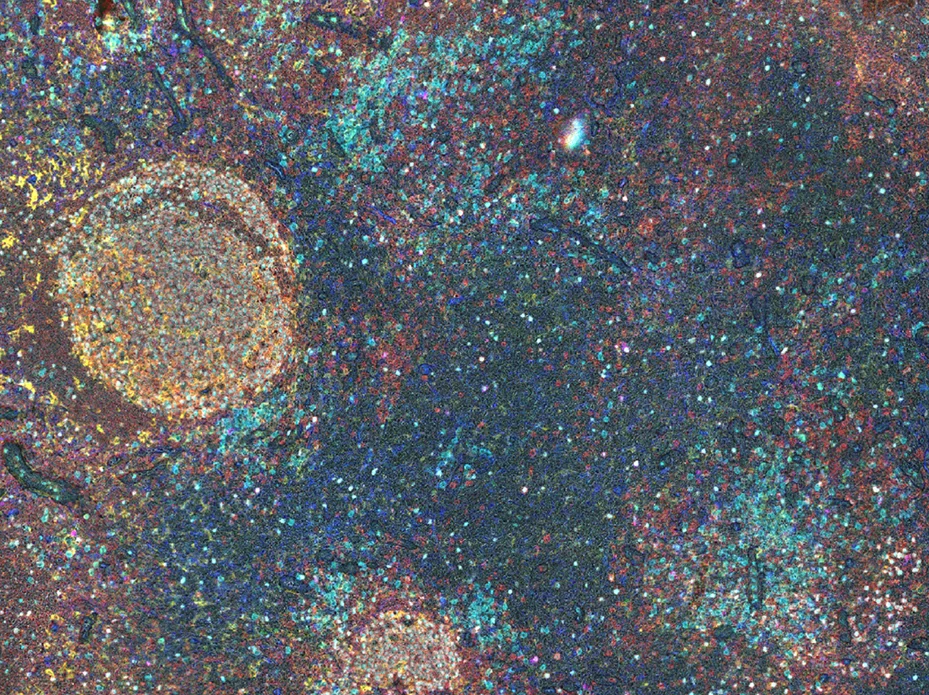
Webinar: High-plex immunohistochemistry - Spatial biology links patient survival with lymph node B cell responses in head & neck cancer
Learn more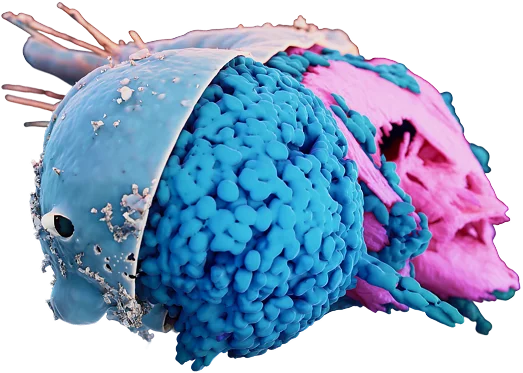
Frequently asked questions
Everything you need to know about the product and billing.
